Manual
Data generation
Here we developed an integrative strategy to predict proteome-wide PPIs between human and HSV-1 strain KOS. All the proteins of the HSV-1 strain KOS were downloaded from GenBank (https://www.ncbi.nlm.nih.gov/nuccore/952947517/). A total number of 20,412 reviewed human proteins used for prediction was downloaded from the UniProt database. We store the predicted PPIs with the integrated predicted score greater than 0.5 in this database, which covers 10,432 PPIs between 4,546 human proteins and 72 HSV-1 proteins.
Interface construction
Network: cytoscape.js
PHP, HTML, CSS
How to download?
You can click "download" at the top of the page to enter the download page.
How to use the search function?
You can search for human or viral proteins of interest to view their predicted PPIs. After selecting the type of protein name entered, that is, gene name, UniProt ID, protein name or GenBank ID, you can type the corresponding name of the protein, or click the blank text box and select from the pop-up drop-down list. You also can click the example to search.
The schematic diagram is as follows:
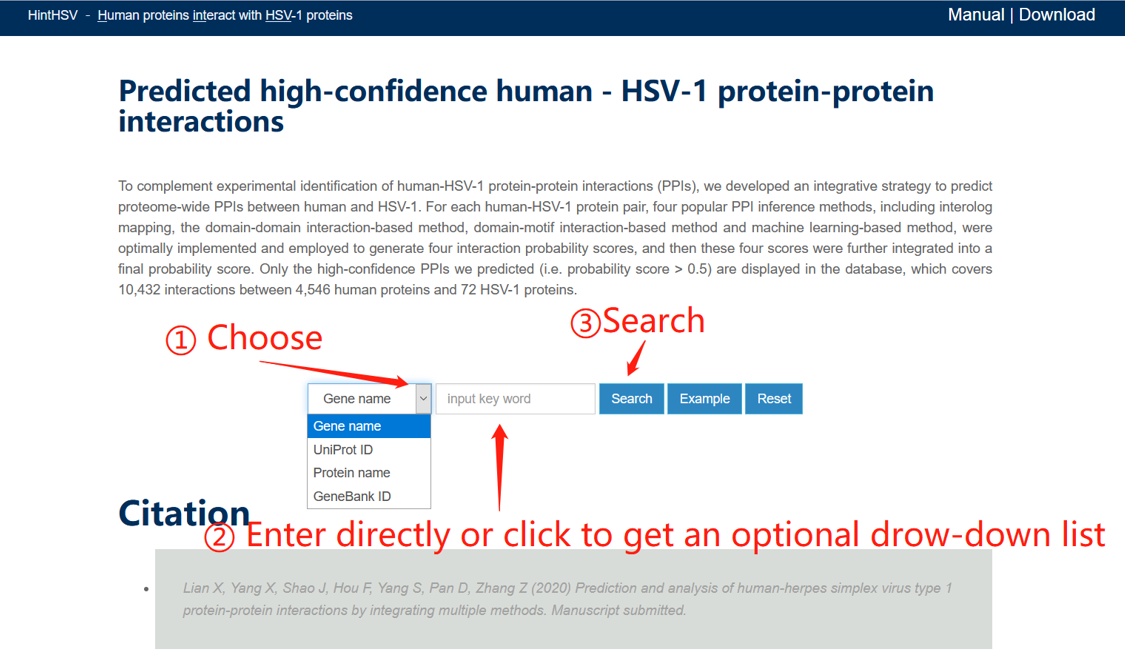
The search results page for the example (Gene name, UL6) is shown below:
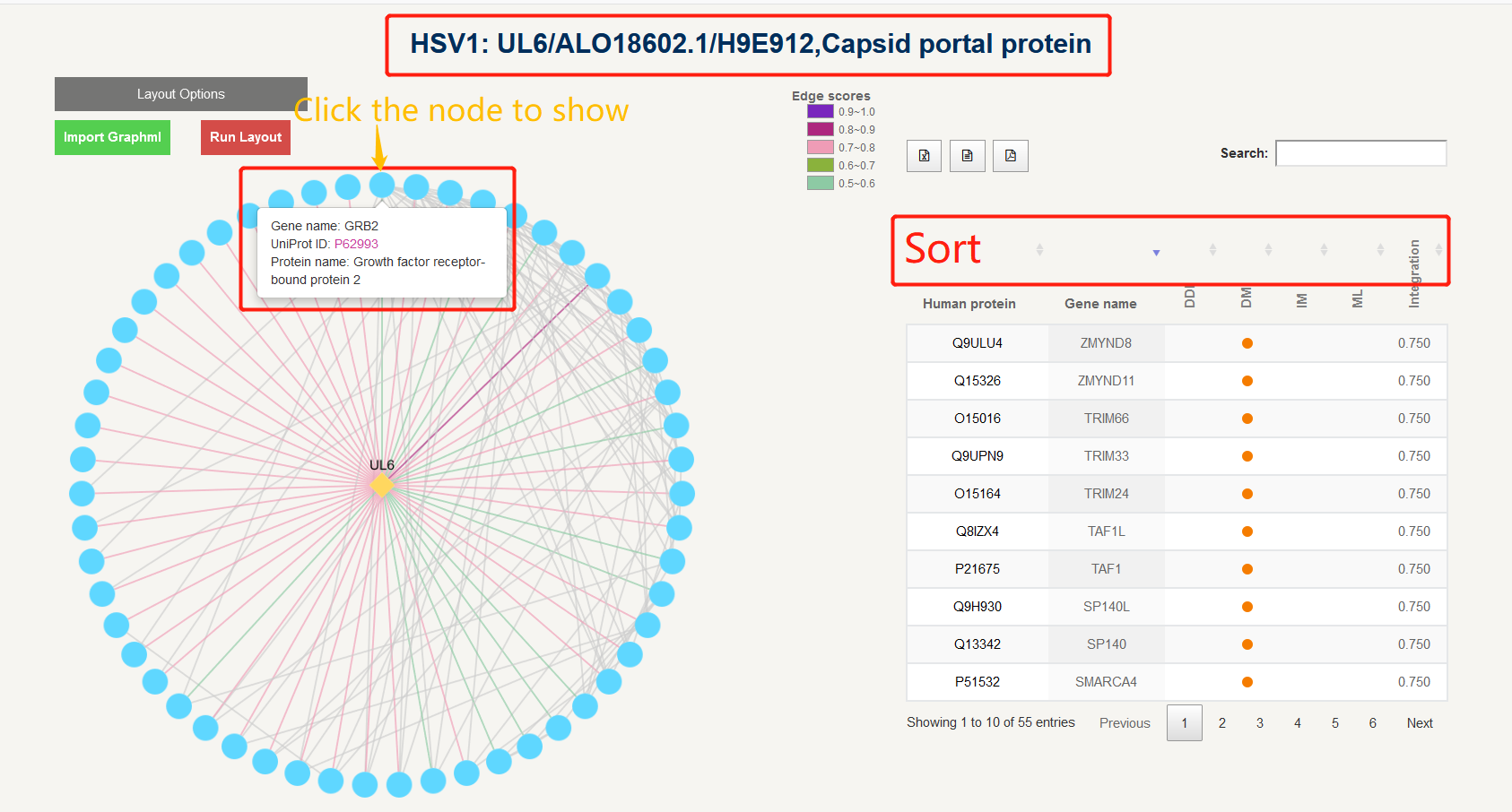
Citation
-
Lian X, Yang X, Shao J, Hou F, Yang S, Pan D, Zhang Z (2020) Prediction and analysis of human-herpes simplex virus type 1 protein-protein interactions by integrating multiple methods. Manuscript submitted.